Intro to the tidyverse
The intent of this chapter is to get you familiarized with the process of going from data import to visualization. This section will walk you through the core concepts in the packages dplyr and ggplot2, which are parts of the tidyverse. The tidyverse is a collection of R packages developed by RStudio’s chief scientist Hadley Wickham. These packages work well together as part of larger data analysis pipeline. You will gain a tremendous level of experience and repetitions with these packages, which will solidify your knowledge. The later chapters will solidify your understanding of all concepts in this part.
Theory Input
Packages
You know what functions are and how to write your own functions. You’re not the only person writing your own functions with R. Many people design tools that can help people analyze data. They provide those functions and objects for free in wrapped up packages. You only have to download and install them. Each R package is hosted at http://cran.r-project.org, the same website that hosts R (often interesting new R packages are only available on GitHub, because submitting to CRAN is a lot more work than just providing a version on github). However, you don’t need to visit the website (or github) to download an R package. You can download packages straight from R’s command line.
Video instructions to install R packages
These are the basic commands you will need for installing and loading packages in your current R session:
# install package from CRAN
install.packages("packagename")
# load the package to use in the current R session
library("packagename")
# use a particular function within a package without loading the package
packagename::function_name()Downlaod the following file and place in your getting_started folder. Try running the commands to install some of the packages we are going to use throughout this class.
Windows Users: You will get a warning if Rtools is not installed. See the last session for install instructions.
If you get a message saying “Do you want to install from source the packages which need compilation?", selecting “No” will revert to the latest compiled version which may not be the most recent on CRAN (the respository that people and organizations store open source packages on). Selecting “Yes” will download the latest, and use your machine to do the compiling. This may take longer.
If you get errors do the following:
- Try installing one package at a time. Find which package has the error and isolate it. Then proceed to step 2.
- Restart R by exiting RStudio and reopening. Retry installing the problem package. If this does not work, proceed to step 3.
- Google “R” + “install.packages” + the error message. Chances are 99% likely that someone else has experienced an error like yours.
If otherwise unsuccessful, contact me and I will do my best to troubleshoot.
Pipes
%>% throughout this class. The “pipe” is from the magrittr package (included in the tidyverse). The point of the pipe is to help you write code in a way that is easier to read and understand. It makes your code more readable by structuring sequences of data operations left-to-right (as opposed to from the inside and out). The pipe makes your code read more like a sentence, branching from left to right. You can read it as a series of imperative statements: group, then summarise, then filter. As suggested by this reading, a good way to pronounce %>% when reading code is “then”. Mathematically it can be expressed like the following:x %>% fis equivalent tof(x)x %>% f(y)is equivalent tof(x, y)x %>% f %>% g %>% his equivalent toh(g(f(x)))
Instead of writing this:
data <- iris
data <- head(data, n=3)you can write the code like this:
# The easiest way to get magrittr is to load the whole tidyverse:
library("tidyverse")
# Alternatively, load just magrittr:
library("magrittr")
iris %>% head(n=3)When coupling several function calls with the pipe-operator, the benefit will become more apparent. Consider this pseudo example:
the_data <-
read.csv('/path/to/data/file.csv') %>%
subset(variable_a > x) %>%
transform(variable_c = variable_a/variable_b) %>%
head(100)Tibbles
tibbles instead of R’s traditional data.frame. Tibbles are data frames, but they tweak some older behaviours to make life a little easier. R is an old language, and some things that were useful 10 or 20 years ago now get in your way. In most places, the term tibble and data frame will be used interchangeably.- tibble is one of the unifying features of tidyverse,
- it is a better data.frame realization,
- objects of the class data.frame can be coerced to tibble using
as_tibble()
When you print a tibble, the output shows:
- all columns that fit the screen
- first 10 rows
- data type for each column
A tibble is being constucted like this:
tibble(
x = 1:50,
y = runif(50),
z = x + y^2,
outcome = rnorm(50)
)Check yourself how dataframes and tibbles print differently (you can use for an example the built in dataframe cars):
class(cars)
## "data.frame"
cars_tbl <- as_tibble(cars)
class(cars_tbl)
## "tbl_df" "tbl" "data.frame"
There are multiple ways of subsetting (retrieving just parts of the date) tibbles:
# This way applies to dataframes and tibbles
vehicles <- as_tibble(cars[1:5,])
vehicles[['speed']]
vehicles[[1]]
vehicles$speed
# Using placeholders with the pipe
vehicles %>% .$dist
vehicles %>% .[['dist']]
vehicles %>% .[[2]]Import
Before you can manipulate data with R, you need to import the data into R’s memory, or build a connection to the data that R can use to access the data remotely.
How you import your data will depend on the format of the data. The most common way to store small data sets is as a plain text file. Data may also be stored in a proprietary format associated with a specific piece of software, such as SAS, SPSS, or Microsoft Excel. Data used on the internet is often stored as a JSON or XML file. Large data sets may be stored in a database or a distributed storage system.
When you import data into R, R stores the data in your computer’s RAM while you manipulate it. This creates a size limitation: truly big data sets should be stored outside of R in a database or distributed storage system. You can then create a connection to the system that R can use to access the data without bringing the data into your computer’s RAM.
The tidyverse offers the following packages for importing data:
readrfor reading flat files like .csv filesreadxlfor .xls and .xlsx sheets.havenfor SPSS, Stata, and SAS data.googledriveto interact with files on Google Drive from R.
There are a handful of other packages that are not in the tidyverse, but are tidyverse-adjacent. They are very useful for importing data from other sources (we will use them in the next session):
- jsonlite for JSON.
- xml2 for XML.
- httr for web APIs.
- rvest for web scraping.
- DBI for relational databases. To connect to a specific database, you’ll need to pair DBI with a specific backend like RSQLite, RPostgres, or odbc. Learn more at https://db.rstudio.com.
Example
# Loading data (can also be achieved by clicking on "Import Dataset > From Text (readr)" in the upper right corner)
library(readr)
dataset_tbl <- read_csv("data.csv")
# In case of possible parsing errors, analyze them with problems()
readr::problems(dataset_tbl)
# Writing data
write_csv(dataset_tbl, "data.csv")
# Saving in csv (or tsv) does mean you loose information about the type of data in particular columns. You can avoid this by using write_rds() and read_rds() to read/write objects in R binary rds format.
write_rds(dataset_tbl, "data.rds")Tidy
Reshaping your data with tidyr
The Concept of Tidy Data
- each and every observation is represented as exactly one row,
- each and every variable is represented by exactly one column,
- thus each data table cell contains only one value.

Usually data are untidy in only one way. However, if you are unlucky, they are really untidy and thus a pain to work with…
Exercise
Which of these data are tidy?
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| Species | variable | value |
|---|---|---|
| setosa | Sepal.Length | 5.1 |
| setosa | Sepal.Length | 4.9 |
| setosa | Sepal.Length | 4.7 |
| Sepal.Length | 5.1 | 4.9 | 4.7 | 4.6 |
|---|---|---|---|---|
| Sepal.Width | 3.5 | 3.0 | 3.2 | 3.1 |
| Petal.Length | 1.4 | 1.4 | 1.3 | 1.5 |
| Petal.Width | 0.2 | 0.2 | 0.2 | 0.2 |
| Species | setosa | setosa | setosa | setosa |
| Sepal.L.W | Petal.L.W | Species |
|---|---|---|
| 5.1/3.5 | 1.4/0.2 | setosa |
| 4.9/3 | 1.4/0.2 | setosa |
| 4.7/3.2 | 1.3/0.2 | setosa |
| --- | --- | --- |
Solution
To illustrate how we can make data tidy easily, we are using modified variants of the diamonds dataset from the ggplot2 package. Try to make the data tidy by looking at the corresponding help pages of each provided function.
- Tidying data with
pivot_longer(): Use, if some of your column names are actually values of a variable. Alternatively you can usegather(), but it is recommended to usepivot_longer(), because the other function is no longer being maintained.
library(tidyverse)
diamonds2 <- readRDS("diamonds2.rds")
diamonds2 %>% head(n = 5)
## # A tibble: 5 x 3
## cut `2008` `2009`
## <chr> <dbl> <dbl>
## 1 Ideal 326 332
## 2 Premium 326 332
## 3 Good 237 333
## 4 Premium 334 340
## 5 Good 335 341Solution
diamonds2 %>%
pivot_longer(cols = c("2008", "2009"),
names_to = 'year',
values_to = 'price') %>%
head(n = 5)
## # A tibble: 5 x 3
## cut year price
## <chr> <chr> <dbl>
## 1 Ideal 2008 326
## 2 Ideal 2009 332
## 3 Premium 2008 326
## 4 Premium 2009 332
## 5 Good 2008 237The wide (original) format might be good for business reports, but it is not good for doing analyses. If we wanted to predict the price based on the other data (with a simple linear regression), we need the long format:
model <- lm(price ~ ., data = diamonds2_long)
model
## Call:
## lm(formula = price ~ ., data = diamonds2_long)
## Coefficients:
## (Intercept) cutIdeal cutPremium year2009
## 237 89 89 6- Tidying data with
pivot_wider(): Use, if some of your observations are scattered across many rows. Alternatively you can usespread(), but it is recommended to usepivot_wider(), because the other function is no longer being maintained.
diamonds3 <- readRDS("diamonds3.rds")
diamonds3 %>% head(n = 5)
## # A tibble: 5 x 5
## cut price clarity dimension measurement
## <ord> <dbl> <ord> <chr> <dbl>
## 1 Ideal 326 SI2 x 3.95
## 2 Premium 326 SI1 x 3.89
## 3 Good 327 VS1 x 4.05
## 4 Ideal 326 SI2 y 3.98
## 5 Premium 326 SI1 y 3.84Solution
diamonds3 %>%
pivot_wider(names_from = "dimension",
values_from = "measurement") %>%
head(n = 5)
## # A tibble: 3 x 6
## cut price clarity x y z
## <ord> <dbl> <ord> <dbl> <dbl> <dbl>
## 1 Ideal 326 SI2 3.95 3.98 2.43
## 2 Premium 326 SI1 3.89 3.84 2.31
## 3 Good 327 VS1 4.05 4.07 2.31- Tidying data with
separate(): If some of your columns contain more than one value, use separate:
diamonds4 <- readRDS("diamonds4.rds")
diamonds4
## # A tibble: 5 x 4
## cut price clarity dim
## <ord> <dbl> <ord> <chr>
## 1 Ideal 326 SI2 3.95/3.98/2.43
## 2 Premium 326 SI1 3.89/3.84/2.31
## 3 Good 327 VS1 4.05/4.07/2.31
## 4 Premium 334 VS2 4.2/4.23/2.63
## 5 Good 335 SI2 4.34/4.35/2.75Solution
diamonds4 %>%
separate(col = dim,
into = c("x", "y", "z"),
sep = "/",
convert = T)
## # A tibble: 5 x 6
## cut price clarity x y z
## <ord> <dbl> <ord> <dbl> <dbl> <dbl>
## 1 Ideal 326 SI2 3.95 3.98 2.43
## 2 Premium 326 SI1 3.89 3.84 2.31
## 3 Good 327 VS1 4.05 4.07 2.31
## 4 Premium 334 VS2 4.2 4.23 2.63
## 5 Good 335 SI2 4.34 4.35 2.75- Tidying data with
unite(): use to paste together multiple columns into one:
diamonds5 <- readRDS("diamonds5.rds")
diamonds5
## # A tibble: 5 x 7
## cut price clarity_prefix clarity_suffix x y z
## <ord> <dbl> <chr> <chr> <dbl> <dbl> <dbl>
## 1 Ideal 326 SI 2 3.95 3.98 2.43
## 2 Premium 326 SI 1 3.89 3.84 2.31
## 3 Good 327 VS 1 4.05 4.07 2.31
## 4 Premium 334 VS 2 4.2 4.23 2.63
## 5 Good 335 SI 2 4.34 4.35 2.75Solution
diamonds5 %>%
unite(clarity, clarity_prefix, clarity_suffix, sep = '')
## # A tibble: 5 x 6
## cut price clarity x y z
## <ord> <dbl> <ord> <dbl> <dbl> <dbl>
## 1 Ideal 326 SI2 3.95 3.98 2.43
## 2 Premium 326 SI1 3.89 3.84 2.31
## 3 Good 327 VS1 4.05 4.07 2.31
## 4 Premium 334 VS2 4.2 4.23 2.63
## 5 Good 335 SI2 4.34 4.35 2.75Transform
Often you’ll need to create some new variables or summaries, or maybe you just want to rename the variables or reorder the observations in order to make the data a little easier to work with. dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges. The following five key dplyr functions allow you to solve the vast majority of your data manipulation challenges:
filter()picks cases based on their values (formula based filtering). Useslice()for filtering with row numbers. So both can be used for selecting the relevant rows.
library(ggplot2) # To load the diamonds dataset
library(dplyr)
diamonds %>%
filter(cut == 'Ideal' | cut == 'Premium', carat >= 0.23) %>%
head(5)
## # A tibble: 5 x 10
## carat cut color clarity depth table price x y z
##
## 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
## 2 0.290 Premium I VS2 62.4 58 334 4.2 4.23 2.63
## 3 0.23 Ideal J VS1 62.8 56 340 3.93 3.9 2.46
## 4 0.31 Ideal J SI2 62.2 54 344 4.35 4.37 2.71
## 5 0.32 Premium E I1 60.9 58 345 4.38 4.42 2.68
diamonds %>%
filter(cut == 'Ideal' | cut == 'Premium', carat >= 0.23) %>%
slice(3:4)
## # A tibble: 2 x 10
## carat cut color clarity depth table price x y z
##
## 1 0.23 Ideal J VS1 62.8 56 340 3.93 3.9 2.46
## 2 0.31 Ideal J SI2 62.2 54 344 4.35 4.37 2.71 | Operator | Description |
|---|---|
| < | less than |
| <= | less than or equal to |
| > | greater than |
| >= | greater than or equal to |
| == | exactly equal to |
| != | not equal to |
| !x | Not x. Gives the opposite logical value. |
| x | y | x OR y |
| x & y | x AND y |
arrange()changes the ordering of the rows
diamonds %>%
arrange(cut, carat, desc(price))
# A tibble: 53,940 x 10
carat cut color clarity depth table price x y z
1 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49
2 0.23 Fair G VVS2 61.4 66 369 3.87 3.91 2.39
3 0.25 Fair F SI2 54.4 64 1013 4.3 4.23 2.32
4 0.25 Fair D VS1 61.2 55 563 4.09 4.11 2.51
5 0.25 Fair E VS1 55.2 64 361 4.21 4.23 2.33
6 0.27 Fair E VS1 66.4 58 371 3.99 4.02 2.66
7 0.290 Fair F SI1 55.8 60 1776 4.48 4.41 2.48
8 0.290 Fair D VS2 64.7 62 592 4.14 4.11 2.67
9 0.3 Fair D IF 60.5 57 1208 4.47 4.35 2.67
10 0.3 Fair E VVS2 51 67 945 4.67 4.62 2.37
# … with 53,930 more rows The NAs always end up at the end of the rearranged tibble.
select()picks (or removes) variables based on their names
diamonds %>%
select(color, clarity, x:z) %>%
head(n = 5)
## # A tibble: 5 x 5
## color clarity x y z
##
## 1 E SI2 3.95 3.98 2.43
## 2 E SI1 3.89 3.84 2.31
## 3 E VS1 4.05 4.07 2.31
## 4 I VS2 4.2 4.23 2.63
## 5 J SI2 4.34 4.35 2.75 Exclusive select:
diamonds %>%
select(-(x:z)) %>%
head(n = 5)
## # A tibble: 5 x 7
## carat cut color clarity depth table price
##
## 1 0.23 Ideal E SI2 61.5 55 326
## 2 0.21 Premium E SI1 59.8 61 326
## 3 0.23 Good E VS1 56.9 65 327
## 4 0.290 Premium I VS2 62.4 58 334
## 5 0.31 Good J SI2 63.3 58 335 Select helpers
starts_with()/ends_with(): helper that selects every column tht starts with a prefix or ends with a suffixcontains(): A select helper that selects any column containing a string of text.everything(): a select() helper that selects every column that has not already been selected. Good for reordering.
diamonds %>%
select(x:z, everything()) %>%
head(n = 5)
## # A tibble: 5 x 10
## x y z carat cut color clarity depth table price
##
## 1 3.95 3.98 2.43 0.23 Ideal E SI2 61.5 55 326
## 2 3.89 3.84 2.31 0.21 Premium E SI1 59.8 61 326
## 3 4.05 4.07 2.31 0.23 Good E VS1 56.9 65 327
## 4 4.2 4.23 2.63 0.290 Premium I VS2 62.4 58 334
## 5 4.34 4.35 2.75 0.31 Good J SI2 63.3 58 335 rename()changes the name of a column.
diamonds %>%
rename(var_x = x) %>%
head(n = 5)
## # A tibble: 5 x 10
## carat cut color clarity depth table price var_x y z
##
## 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
## 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
## 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
## 4 0.290 Premium I VS2 62.4 58 334 4.2 4.23 2.63
## 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75 mutate()adds new variables that are functions of existing variables and preserves existing ones.transmute()adds new variables and drops existing ones.
diamonds %>%
mutate(p = x + z, q = p + y) %>%
select(-(depth:price)) %>%
head(n = 5)
## # A tibble: 5 x 9
## carat cut color clarity x y z p q
##
## 1 0.23 Ideal E SI2 3.95 3.98 2.43 6.38 10.4
## 2 0.21 Premium E SI1 3.89 3.84 2.31 6.2 10.0
## 3 0.23 Good E VS1 4.05 4.07 2.31 6.36 10.4
## 4 0.290 Premium I VS2 4.2 4.23 2.63 6.83 11.1
## 5 0.31 Good J SI2 4.34 4.35 2.75 7.09 11.4 diamonds %>%
transmute(carat, cut, sum = x + y + z) %>%
head(n = 5)
## # A tibble: 5 x 3
## carat cut sum
##
## 1 0.23 Ideal 10.4
## 2 0.21 Premium 10.0
## 3 0.23 Good 10.4
## 4 0.290 Premium 11.1
## 5 0.31 Good 11.4 bind_cols()andbind_rows(): binds two tibbles column-wise or row-wise.group_by()andsummarize()reduces multiple values down to a single summary
diamonds %>%
group_by(cut) %>%
summarize(max_price = max(price),
mean_price = mean(price),
min_price = min(price))
## # A tibble: 5 x 4
## cut max_price mean_price min_price
##
## 1 Fair 18574 4359. 337
## 2 Good 18788 3929. 327
## 3 Very Good 18818 3982. 336
## 4 Premium 18823 4584. 326
## 5 Ideal 18806 3458. 326 glimpse()can be used to see the columns of the dataset and display some portion of the data with respect to each attribute that can fit on a single line. You can apply this function to get a glimpse of your dataset. It is similar to the base functionstr().
glimpse(diamonds)
## Rows: 53,940
## Columns: 10
## $ carat <dbl> 0.23, 0.21, 0.23, 0.29, 0.31, 0.24, 0.24, 0.26, 0.22, 0.23, …
## $ cut <ord> Ideal, Premium, Good, Premium, Good, Very Good, Very Good, …
## $ color <ord> E, E, E, I, J, J, I, H, E, H, J, J, F, J, E, E, I, J, J, J, …
## $ clarity <ord> SI2, SI1, VS1, VS2, SI2, VVS2, VVS1, SI1, VS2, VS1, SI1, VS1, …
## $ depth <dbl> 61.5, 59.8, 56.9, 62.4, 63.3, 62.8, 62.3, 61.9, 65.1, 59.4, …
## $ table <dbl> 55, 61, 65, 58, 58, 57, 57, 55, 61, 61, 55, 56, 61, 54, 62, …
## $ price <int> 326, 326, 327, 334, 335, 336, 336, 337, 337, 338, 339, 340, 3…
## $ x <dbl> 3.95, 3.89, 4.05, 4.20, 4.34, 3.94, 3.95, 4.07, 3.87, 4.00, …
## $ y <dbl> 3.98, 3.84, 4.07, 4.23, 4.35, 3.96, 3.98, 4.11, 3.78, 4.05, …
## $ z <dbl> 2.43, 2.31, 2.31, 2.63, 2.75, 2.48, 2.47, 2.53, 2.49, 2.39, …Data types and data structures
Everything in R is an object. R has 6 basic data types. U can use typeof() to asses the data type or mode of an object.
| data type | example |
|---|---|
| character | |
| numeric | |
| integer | |
| logical | |
| complex | will not be discussed in this class |
| raw | will not be discussed in this class |
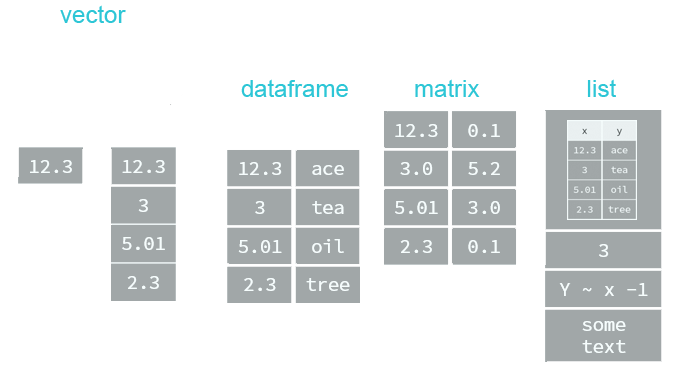
" and single quotes ' can be used interchangeably (most of time) to define a character string, but double quotes are preferred.Most datasets we work with consist of batches of values such as a table of temperature values or a list of survey results. These batches are stored in R in one of several data structures. R has many data structures. These include
- atomic vector
- list
- matrix
- data frame
- factors
We will discuss those as soon as we will be working with them.
At the end of this session we will be dealing with dates. Date values are stored as numbers. But to be properly interpreted as a date object in R, their attribute must be explicitly defined as a date. Attributes are part of the object. These include:
- names
- dimnames
- dim
- class
- attributes (contain metadata)
R provides many facilities to convert and manipulate dates and times, but a package called lubridate makes working with dates/times much easier.
- Easy and fast parsing of date-times:
ymd(),ymd_hms(),dmy(),dmy_hms,mdy(), …
library(lubridate)
ymd(20101215)
## "2010-12-15"
mdy("4/1/17")
## "2017-04-01"- Simple functions to get and set components of a date-time, such as
year(),month(),mday(),hour(),minute()andsecond():
bday <- dmy("14/10/1979")
month(bday)
## 10
year(bday)
## 1979Business case
Goal & Data
Goal
You are a data scientist now. Your job is to study the products, look for opportunities to sell new products, better serve the customer and better market the products. All of that is supposed to be justified by data.
In this session you are about to get your hands into R with a real world situation. The goal is to analyze the sales of bikes sold through bike stores in germany. The bike models correspond to the models of the manufacturer Canyon (in the next session you will see how to gather this data), but the sales and store data are made up.
- Sales by year
- Sales by year and product family
We are going to do that by importing, wrangling and visualizing of the provided data.
Data
The bike sales data is divided in multiple datasets for better understanding and organization. When working with transactional data, Entity-relationship diagrams (ERD) are used for describing and defining the data models (see example below). It illustrates the logical structure of the databases (see
ER-diagram-symbols-and-meaning for further information). Please refer to the following data schema when working with the sales data. It shows with which key column we can combine the single databases. Example: To see which items are included in an order, you have to combine Order Lines with Bikes via the columns product.id and bike.id.

The dataset has information of ~15k orders from 2015 to 2019 made from multiple bike stores in germany. Its features allows viewing an order from multiple dimensions: from price to customer location, product attributes and many more.
The three tables contain the following information (excerpt):
1: Table bikes:
| bike.id | model | model.year | frame.material | weight | price | category | gender | url |
|---|---|---|---|---|---|---|---|---|
| 2875 | Aeroad CF SL Disc 8.0 Di2 | 2020 | carbon | 7.60 | 4579 | Road - Race - Aeroad | unisex | https://www.canyon.com/en-de/road-bikes/race-bikes/aeroad/aeroad-cf-sl-disc-8.0-di2/2875.html |
| … | … | … | … | … | … | … | … | … |
bike.id: Unique product identifiermodel: Product namemodel.year: Release date of the bike modelframe.material: Aluminium or carbonweight: Product weight measured in kilograms.price: Item pricecategory: Bike family, bike category and bike ride style (separated by “-")gender: Unisex or femaleurl: Link to the product in the online store of Canyon.
2: Table bikeshops:
| bikeshop.id | name | location | lat | lng | ||
|---|---|---|---|---|---|---|
| 6 | fahrschneller | Stuttgart, Baden-Württemberg | 48.8 | 9.18 | ||
| … | … | … | … | … |
bikeshop.id: Seller unique identifiername: Name of the bikeshoplocation: City name and state of the bikeshop (separated by “,")lat: Latitudelng: Longitude
3: Table Order Lines:
| ___ | order.id | order.line | order.date | customer.id | product.id | quantity |
|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 2015-01-07 | 2 | 2681 | 1 |
| … | … | … | … | … | … | … |
___: Rownumberorder.id: Unique identifier of the order.order.line: Sequential number identifying number of different items included in the same order.order.date: Shows the purchase timestamp.customer.id: Key to the bikeshop dataset.product.id: Key to the bikes datasetquantity: Quantity of the ordered bikes per order line.
Analysis with R
First steps
You have downloaded the data already in the last session. Let’s start by creating a script file. You can download the following template and add it to your folder 01_getting_started:
---- (four dashes) at the end of a comment.As you can see the template has seven sections. In the following we are going to populate them step by step in order to conduct our analysis.
Session and then click Restart R1. Libraries
You can just load the tidyverse library since it attaches all the packages we will need for this analysis. For the purpose of learning, the single packages, that we need, are listed as well.
# 1.0 Load libraries ----
library(tidyverse)
# library(tibble) --> is a modern re-imagining of the data frame
# library(readr) --> provides a fast and friendly way to read rectangular data like csv
# library(dplyr) --> provides a grammar of data manipulation
# library(magrittr) --> offers a set of operators which make your code more readable (pipe operator)
# library(tidyr) --> provides a set of functions that help you get to tidy data
# library(stringr) --> provides a cohesive set of functions designed to make working with strings as easy as possible
# library(ggplot2) --> graphics
## ── Attaching packages ──────────────────────────────── tidyverse 1.3.0 ──
## ✓ ggplot2 3.3.0 ✓ purrr 0.3.4
## ✓ tibble 3.0.1 ✓ dplyr 0.8.5
## ✓ tidyr 1.0.2 ✓ stringr 1.4.0
## ✓ readr 1.3.1 ✓ forcats 0.5.0
## ── Conflicts ───────────────────────────────────────── tidyverse_conflicts() ──
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()
# Excel Files
library(readxl)2. Import
The files are located at /00_data/01_bike_sales/01_raw_data/. To read tha data into R we are going to use the read_excel() function from the readxl package. Take a look at the help site to figure out how to use it. Also think about which data we need for our analysis. You can ignore the default arguments (the arguments which equal already a value) for now. Don’t forget to store the data into a named variable.
?read_excelWe need:
- orderlines (for years)
- bikes (for price and for the product families)
# 2.0 Importing Files ----
# A good convention is to use the file name and suffix it with tbl for the data structure tibble
bikes_tbl <- read_excel(path = "00_data/01_bike_sales/01_raw_data/bikes.xlsx")
orderlines_tbl <- read_excel("00_data/01_bike_sales/01_raw_data/orderlines.xlsx")
# Not necessary for this analysis, but for the sake of completeness
bikeshops_tbl <- read_excel("00_data/01_bike_sales/01_raw_data/bikeshops.xlsx")
In your environment you should see now 3 loaded tables. You can click on them to take a look at the data.
3. Examine
Use different methods to take a look and get a feel for the data.
# 3.0 Examining Data ----
# Method 1: Print it to the console
orderlines_tbl
# A tibble: 15,644 x 7
## ...1 order.id order.line order.date customer.id product.id quantity
## <chr> <dbl> <dbl> <dttm> <dbl> <dbl> <dbl>
## 1 1 1 1 2015-01-07 00:00:00 2 2681 1
## 2 2 1 2 2015-01-07 00:00:00 2 2411 1
## 3 3 2 1 2015-01-10 00:00:00 10 2629 1
## 4 4 2 2 2015-01-10 00:00:00 10 2137 1
## 5 5 3 1 2015-01-10 00:00:00 6 2367 1
## 6 6 3 2 2015-01-10 00:00:00 6 1973 1
## 7 7 3 3 2015-01-10 00:00:00 6 2422 1
## 8 8 3 4 2015-01-10 00:00:00 6 2655 1
## 9 9 3 5 2015-01-10 00:00:00 6 2247 1
## 10 10 4 1 2015-01-11 00:00:00 22 2408 1
## # … with 15,634 more rows
# Method 2: Clicking on the file in the environment tab (or run View(orderlines_tbl)) There you can play around with the filter.
# Method 3: glimpse() function. Especially helpful for wide data (data with many columns)
glimpse(orderlines_tbl)
## Rows: 15,644
## Columns: 7
## $ ...1 <chr> "1", "2", "3", "4", "5", "6", "7", "8", "…
## $ order.id <dbl> 1, 1, 2, 2, 3, 3, 3, 3, 3, 4, 5, 5, 5, 5,…
## $ order.line <dbl> 1, 2, 1, 2, 1, 2, 3, 4, 5, 1, 1, 2, 3, 4,…
## $ order.date <dttm> 2015-01-07, 2015-01-07, 2015-01-10, 2015…
## $ customer.id <dbl> 2, 2, 10, 10, 6, 6, 6, 6, 6, 22, 8, 8, 8,…
## $ product.id <dbl> 2681, 2411, 2629, 2137, 2367, 1973, 2422,…
## $ quantity <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1,…Data manipulation
4. Joining data
Take a look at ?left_join(). Start by merging order_items and products and then chain all joins together.
# by automatically detecting a common column, if any ...
left_join(orderlines_tbl, bikes_tbl)
## Error: `by` must be supplied when `x` and `y` have no common variables.
# If the data has no common column name, you can provide each column name in the "by" argument. For example, by = c("a" = "b") will match x.a to y.b. The order of the columns has to match the order of the tibbles).
left_join(orderlines_tbl, bikes_tbl, by = c("product.id" = "bike.id"))
## # A tibble: 15,644 x 15
## ...1 order.id order.line order.date customer.id
## <chr> <dbl> <dbl> <dttm> <dbl>
## 1 1 1 1 2015-01-07 00:00:00 2
## 2 2 1 2 2015-01-07 00:00:00 2
## 3 3 2 1 2015-01-10 00:00:00 10
## 4 4 2 2 2015-01-10 00:00:00 10
## 5 5 3 1 2015-01-10 00:00:00 6
## 6 6 3 2 2015-01-10 00:00:00 6
## 7 7 3 3 2015-01-10 00:00:00 6
## 8 8 3 4 2015-01-10 00:00:00 6
## 9 9 3 5 2015-01-10 00:00:00 6
## 10 10 4 1 2015-01-11 00:00:00 22
## # … with 15,634 more rows, and 10 more variables:
## # product.id <dbl>, quantity <dbl>, model <chr>,
## # model.year <dbl>, frame.material <chr>, weight <dbl>,
## # price <dbl>, category <chr>, gender <chr>, url <chr>
# Chaining commands with the pipe and assigning it to order_items_joined_tbl
bike_orderlines_joined_tbl <- orderlines_tbl %>%
left_join(bikes_tbl, by = c("product.id" = "bike.id")) %>%
left_join(bikeshops_tbl, by = c("customer.id" = "bikeshop.id"))
# Examine the results with glimpse()
bike_orderlines_joined_tbl %>% glimpse()
## Rows: 15,644
## Columns: 19
## $ ...1 <chr> "1", "2", "3", "4", "5", "6", "7", …
## $ order.id <dbl> 1, 1, 2, 2, 3, 3, 3, 3, 3, 4, 5, 5,…
## $ order.line <dbl> 1, 2, 1, 2, 1, 2, 3, 4, 5, 1, 1, 2,…
## $ order.date <dttm> 2015-01-07, 2015-01-07, 2015-01-10…
## $ customer.id <dbl> 2, 2, 10, 10, 6, 6, 6, 6, 6, 22, 8,…
## $ product.id <dbl> 2681, 2411, 2629, 2137, 2367, 1973,…
## $ quantity <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2,…
## $ model <chr> "Spectral CF 7 WMN", "Ultimate CF S…
## $ model.year <dbl> 2021, 2020, 2021, 2019, 2020, 2020,…
## $ frame.material <chr> "carbon", "carbon", "carbon", "carb…
## $ weight <dbl> 13.80, 7.44, 14.06, 8.80, 11.50, 8.…
## $ price <dbl> 3119, 5359, 2729, 1749, 1219, 1359,…
## $ category <chr> "Mountain - Trail - Spectral", "Roa…
## $ gender <chr> "female", "unisex", "unisex", "unis…
## $ url <chr> "https://www.canyon.com/en-de/mount…
## $ name <chr> "AlexandeRad", "AlexandeRad", "WITT…
## $ location <chr> "Hamburg, Hamburg", "Hamburg, Hambu…
## $ lat <dbl> 53.57532, 53.57532, 53.07379, 53.07…
## $ lng <dbl> 10.015340, 10.015340, 8.826754, 8.8…You should have a new variable called bike_orderlines_joined_tbl stored in your Global Environment.
5. Wrangling data
Data manipulation & Cleaning. Usually a data scientist will spend most of his/her time at this section. These are the issues we are facing for our analysis:
- Take a look at the column
category(e.g. by runningbike_orderlines_joined_tbl$category). Those entries seem to have three categories separated by a-. For example there are Mountain - Trail - Spectral and Mountain - Trail - Neuron. You can print all unique entries, that start withMountainwith the following code chunk:
bike_orderlines_joined_tbl %>%
select(category) %>%
filter(str_detect(category, "^Mountain")) %>%
unique()
## # A tibble: 10 x 1
## category
## <chr>
## 1 Mountain - Trail - Spectral
## 2 Mountain - Trail - Neuron
## 3 Mountain - Dirt Jump - Stitched
## 4 Mountain - Enduro - Torque
## 5 Mountain - Trail - Grand Canyon
## 6 Mountain - Cross-Country - Lux
## 7 Mountain - Enduro - Strive
## 8 Mountain - Downhill - Sender
## 9 Mountain - Fat Bikes - Dude
## 10 Mountain - Cross-Country - ExceedWe want to analyze the total sales by product family (= 1st category). To do it, we have to separate the column category in category.1, category.2 and category.3 and add the total price (sales price * quantity) to the data.
To fix those issues we are going to do a series of dplyr operations on the object bike_orderlines_joined_tbl:
# 5.0 Wrangling Data ----
# All actions are chained with the pipe already. You can perform each step separately and use glimpse() or View() to validate your code. Store the result in a variable at the end of the steps.
bike_orderlines_wrangled_tbl <- bike_orderlines_joined_tbl %>%
# 5.1 Separate category name
separate(col = category,
into = c("category.1", "category.2", "category.3"),
sep = " - ") %>%
# 5.2 Add the total price (price * quantity)
# Add a column to a tibble that uses a formula-style calculation of other columns
mutate(total.price = price * quantity) %>%
# 5.3 Optional: Reorganize. Using select to grab or remove unnecessary columns
# 5.3.1 by exact column name
select(-...1, -gender) %>%
# 5.3.2 by a pattern
# You can use the select_helpers to define patterns.
# Type ?ends_with and click on Select helpers in the documentation
select(-ends_with(".id")) %>%
# 5.3.3 Actually we need the column "order.id". Let's bind it back to the data
bind_cols(bike_orderlines_joined_tbl %>% select(order.id)) %>%
# 5.3.4 You can reorder the data by selecting the columns in your desired order.
# You can use select_helpers like contains() or everything()
select(order.id, contains("order"), contains("model"), contains("category"),
price, quantity, total.price,
everything()) %>%
# 5.4 Rename columns because we actually wanted underscores instead of the dots
# (one at the time vs. multiple at once)
rename(bikeshop = name) %>%
set_names(names(.) %>% str_replace_all("\\.", "_"))Explanation of the last step:
names()returns all of the column names of a tibble as a character vector.- The dot
.is used in dplyr pipes%>%to supply the incoming data in another part of the function. The dot enables passing the incoming tibble to multiple spots in the function. str_replace_all()takes a "pattern" argument to find a pattern using Regular Expressions (RegEx) and a "replacement" argument to replace the pattern. Regex is used in programming to match strings. The period.is a special character. It needs to be "escaped" using"\\.". We will use RegEx again in the next session.
Insights & Saving
6. Business Insights
Now that we got the data in a good format, we can start to create business insights for that. We are going to do two analyses. The first is Sales by year and the second is sales by category. This will be a 2 step process for each analysis: Step 1: Manipulate / prepare the data and Step 2: Visualize it (this is one of the most important skills a data scientist needs to know).
Sales by year: Step 1
In the first step we need to do the following:
- Select the columns we need
- Extract the years from the date (we will use the lubridate library)
- Group the data by years and summarize the sales
# 6.0 Business Insights ----
# 6.1 Sales by Year ----
library(lubridate)
# Step 1 - Manipulate
sales_by_year_tbl <- bike_orderlines_wrangled_tbl %>%
# Select columns
select(order_date, total_price) %>%
# Add year column
mutate(year = year(order_date)) %>%
# Grouping by year and summarizing sales
group_by(year) %>%
summarize(sales = sum(total_price)) %>%
# Optional: Add a column that turns the numbers into a currency format
# (makes it in the plot optically more appealing)
# mutate(sales_text = scales::dollar(sales)) <- Works for dollar values
mutate(sales_text = scales::dollar(sales, big.mark = ".",
decimal.mark = ",",
prefix = "",
suffix = " €"))
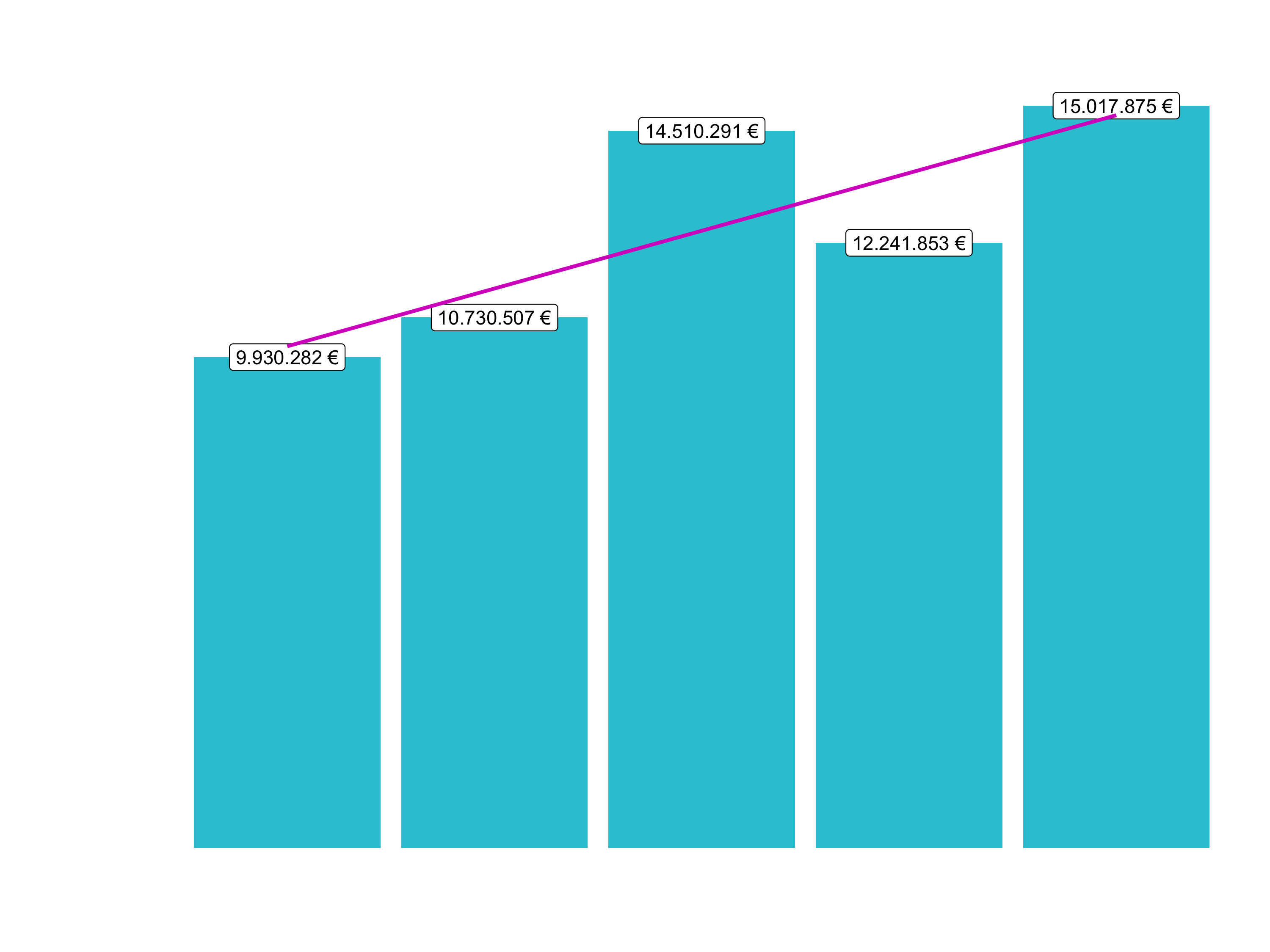
sales_by_year_tbl
## # A tibble: 5 x 3
## year sales sales_text
## <dbl> <dbl> <chr>
## 1 2015 9930282 9.930.282 €
## 2 2016 10730507 10.730.507 €
## 3 2017 14510291 14.510.291 €
## 4 2018 12241853 12.241.853 €
## 5 2019 15017875 15.017.875 €Sales by year: Step 2
Now that we got the data in a good format, we can visualize it with ggplot2. A bar plot is well suited for this case. This section will give you just some exposure to visualization. Session 5 will be completely dedicated to this topic.
- Create a canvas
- Select plotting style (bar plot in this case)
- Format the plot
# 6.1 Sales by Year ----
# Step 2 - Visualize
sales_by_year_tbl %>%
# Setup canvas with the columns year (x-axis) and sales (y-axis)
ggplot(aes(x = year, y = sales)) +
# Geometries
geom_col(fill = "#2DC6D6") + # Use geom_col for a bar plot
geom_label(aes(label = sales_text)) + # Adding labels to the bars
geom_smooth(method = "lm", se = FALSE) + # Adding a trendline
# Formatting
# scale_y_continuous(labels = scales::dollar) + # Change the y-axis.
# Again, we have to adjust it for euro values
scale_y_continuous(labels = scales::dollar_format(big.mark = ".",
decimal.mark = ",",
prefix = "",
suffix = " €")) +
labs(
title = "Revenue by year",
subtitle = "Upward Trend",
x = "", # Override defaults for x and y
y = "Revenue"
)
Revenue by years was a first great vizualisation. But often times, what we want to do is dive into subsets of data. Showing Revenue by year and by category is a good way to extract more details from the existing data.
Sales by category: Step 1
- Select columns
- Group by year and category simultaneously
# 6.2 Sales by Year and Category ----
# Step 1 - Manipulate
sales_by_year_cat_1_tbl <- bike_orderlines_wrangled_tbl %>%
# Select columns and add a year
select(order_date, total_price, category_1) %>%
mutate(year = year(order_date)) %>%
# Group by and summarize year and main catgegory
group_by(year, category_1) %>%
summarise(sales = sum(total_price)) %>%
ungroup() %>%
# Format $ Text
mutate(sales_text = scales::dollar(sales, big.mark = ".",
decimal.mark = ",",
prefix = "",
suffix = " €"))
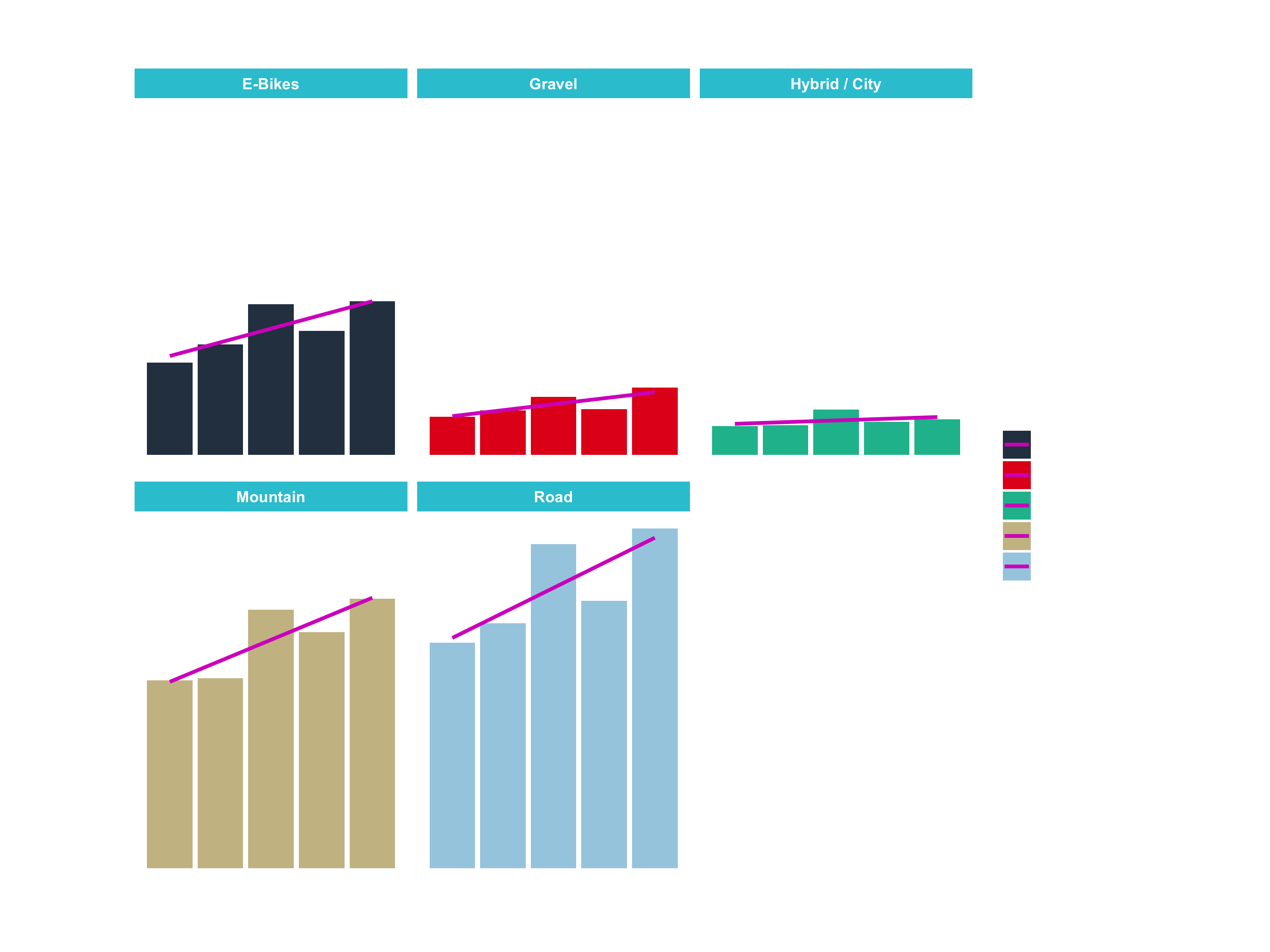
sales_by_year_cat_1_tbl
## # A tibble: 25 x 4
## year category_1 sales sales_text
## <dbl> <chr> <dbl> <chr>
## 1 2015 E-Bikes 1599048 1.599.048 €
## 2 2015 Gravel 663025 663.025 €
## 3 2015 Hybrid / City 502512 502.512 €
## 4 2015 Mountain 3254289 3.254.289 €
## 5 2015 Road 3911408 3.911.408 €
## 6 2016 E-Bikes 1916469 1.916.469 €
## 7 2016 Gravel 768794 768.794 €
## 8 2016 Hybrid / City 512346 512.346 €
## 9 2016 Mountain 3288733 3.288.733 €
## 10 2016 Road 4244165 4.244.165 €
## # … with 15 more rowsSales by category: Step 2
- Create a canvas
- Select plotting style (combination of a facet wrap and bar plots in this case)
# Step 2 - Visualize
sales_by_year_cat_1_tbl %>%
# Set up x, y, fill
ggplot(aes(x = year, y = sales, fill = category_1)) +
# Geometries
geom_col() + # Run up to here to get a stacked bar plot
# Facet
facet_wrap(~ category_1) +
# Formatting
scale_y_continuous(labels = scales::dollar_format(big.mark = ".",
decimal.mark = ",",
prefix = "",
suffix = " €")) +
labs(
title = "Revenue by year and main category",
subtitle = "Each product category has an upward trend",
fill = "Main category" # Changes the legend name
)
Checks at this Point
- Make sure that you have two plots, one for aggregated sales by year and one for sales by year and category_1
- Save your work
7. Writing files
Excel is great when others may want access to your data that are Excel users. For example, many Busienss Intelligence Analysts use Excel not R. CSV is a good option when others may use different languages such as Python, Java or C++. RDS is a format used exclusively by R to save any R object in it’s native format
# 7.0 Writing Files ----
# 7.1 Excel ----
install.packages("writexl")
library("writexl")
bike_orderlines_wrangled_tbl %>%
write_xlsx("00_data/01_bike_sales/02_wrangled_data/bike_orderlines.xlsx")
# 7.2 CSV ----
bike_orderlines_wrangled_tbl %>%
write_csv("00_data/01_bike_sales/02_wrangled_data/bike_orderlines.csv")
# 7.3 RDS ----
bike_orderlines_wrangled_tbl %>%
write_rds("00_data/01_bike_sales/02_wrangled_data/bike_orderlines.rds")Datacamp
Challenge
Your challenges are pretty similar like the analyses we just did:
- Analyze the sales by location (state) with a bar plot. Since
stateandcityare multiple features (variables), they should be split. Which state has the highes revenue? Replace yourbike_orderlines_wrangled_tblobject with the newly wrangled object (with the columnsstateandcity).
Hint: Add + theme(axis.text.x = element_text(angle = 45, hjust = 1)) to your plotting code to rotate your x-axis labels. Probably you have to resize the viewer pane to show the entire plot. For your website, try different values for fig.width and fig.height in your markdown document:
```{r plot, fig.width=10, fig.height=7}
sales_by_loc_tbl %>%
ggplot( ... ) +
...
```
- Analyze the sales by location and year (facet_wrap). Because there are 12 states with bike stores, you should get 12 plots.
Insert your scripts and results into your website. It might be easier to move your entire project folder into your website folder.



