Introduction to R, RStudio IDE & GitHub


This chapter provides a broad overview of the R language that will get you programming right away. The first project will be creating a decision tool used in cost accounting. This project will make it easier to study these things by teaching you the basics of R. The term “R” is used to refer to both the programming language and the software that interprets the scripts written using it. Your first mission is simple: assemble R code that will calculate the ideal quantity of inventory to order for a given product (Economic Order Quantity).
In this project, you will learn how to:
- Use the R and RStudio interfaces
- Run R commands
- Create R objects
- Write your own R functions and scripts
Don’t worry if you’ve never programmed before and it seems like we cover a lot of ground fast. The chapter will teach you everything you need to know and gives you a concise overview of the R language. You will return to many of the concepts we meet here in the next projects, where you will examine the concepts in depth.
Before you can ask your computer to save some numbers, you’ll need to know how to talk to it. RStudio gives you a way to talk to your computer. R gives you a language to speak in. To start you will need to have both R and RStudio installed on your computer before you can use them. R and RStudio are separate downloads and installations. Both are free (under the Affero General Public License (AGPL) v3) and easy to download. R is the underlying statistical computing environment, but using R alone is no fun. RStudio is a graphical integrated development environment (IDE) that makes using R much easier and more interactive. You need to install R before you install RStudio. In the following sections you’ll find information about how to install the language R and the IDE RStudio. You can choose between an interactive and a manual tutorial.
Installing R & RStudio IDE
Interactively
Take the following stepts to get R and the IDERStudio running:
If you are using safari as a browser, you have to open the content in a new window. Go to the following link and follow the instructions to install R and RStudio.
Manually
Download and install R from CRAN, the Comprehensive R Archive Network.
Video instructions to install R (Download link below)
Steps
- Click “Download R for Mac/Windows”
- Download the appropriate file:
- Windows users click Base, and download the installer for the latest R version
- Mac users select the file R-3.X.X.pkg that aligns with your OS version
- Follow the instructions of the installer.
Install RStudio’s IDE
Video instructions to install RStudio (Download link below)
Steps
- Select the install file for your OS.
- Follow the instructions of the installer.
Understanding the RStudio IDE
Let’s start by learning about RStudio, the Integrated Development Environment (IDE) for working with R. We will use RStudio IDE to write code, navigate the files on our computer, inspect the variables we are going to create, and visualize the plots we will generate. RStudio can also be used for other things (e.g., version control, developing packages, etc.) that we will not cover during this class.
One of the advantages of using RStudio is that all the information you need to write code is available in a single window. Additionally, with many shortcuts, autocompletion and highlighting for the major file types you use while developing in R, RStudio will make typing easier and less error-prone. To get started, open RStudio just as you would open any other application on your computer. When you do, a window should appear in your screen like the one shown in the Figure below. RStudio is divided into 4 Panes.
At first startup you will not see an open script as seen in the green frame. You can create one by clicking first on ![]() (new file) and then on
(new file) and then on ![]() (new R script) in the upper left corner. Alternatively you can use the shortcut Shift+⌘+N on macOS or Shift+Ctrl+N on Windows & Linux.
(new R script) in the upper left corner. Alternatively you can use the shortcut Shift+⌘+N on macOS or Shift+Ctrl+N on Windows & Linux.
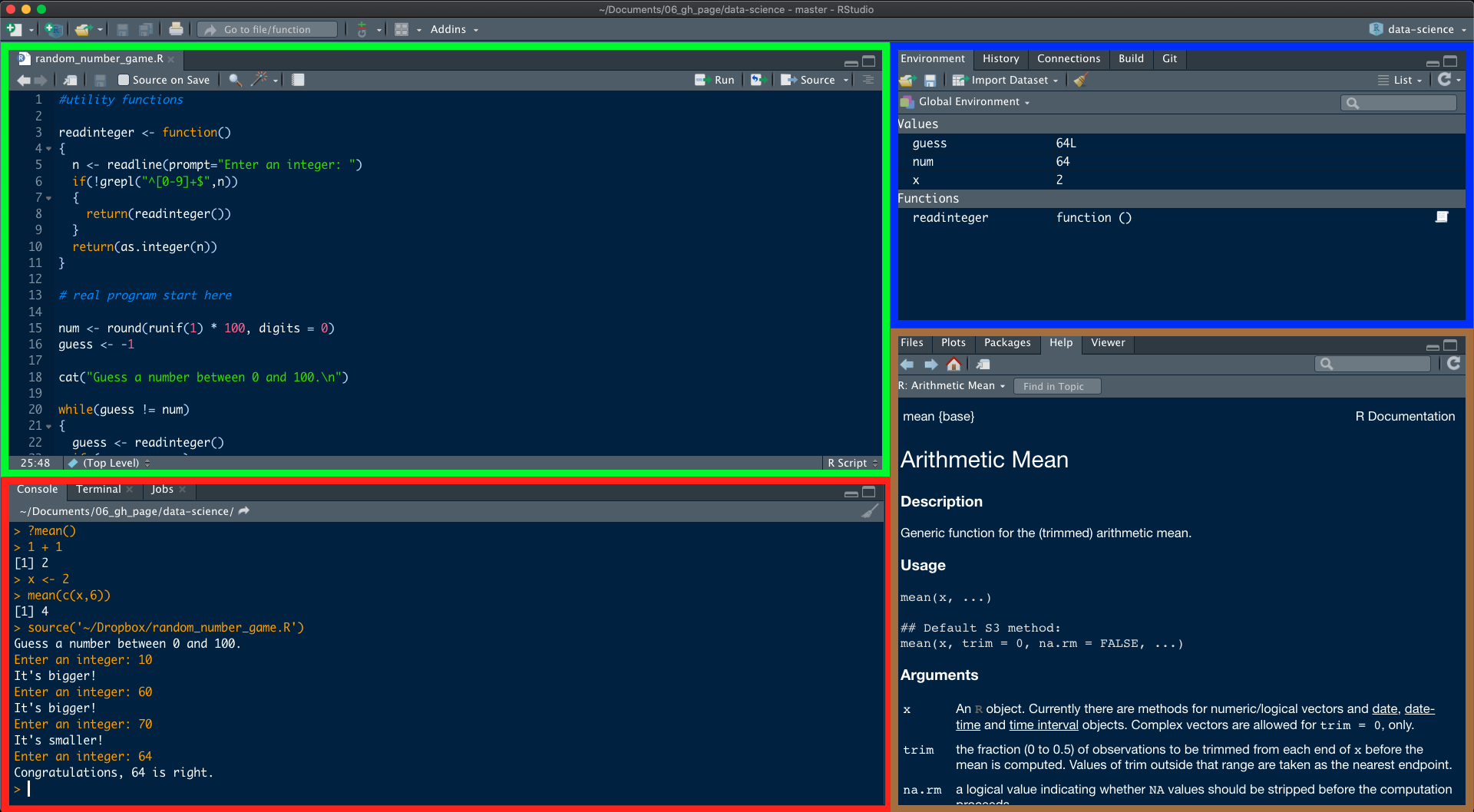
The basis of programming is that we write down instructions for the computer to follow, and then we tell the computer to follow those instructions. We write, or code, instructions in R because it is a common language that both the computer and we can understand. We call the instructions commands and we tell the computer to follow the instructions by executing (also called running) those commands. There are two main ways of interacting with R: by using the console or by using script files.
Console
> prompt for your next command. For example, if you type 10 * 10 and hit Enter, RStudio will display:> 10 * 10
[1] 100
>You’ll notice that a [1] appears next to your result. R is just letting you know that this line begins with the first value in your result. Some commands return more than one value, and their results may fill up multiple lines. For example, the command 1:100 returns 100 integer (numbers that have no digits after the decimal point). It creates a sequence of integers from 1 to 100. Notice that new bracketed numbers appear at the start of the second and third lines of output. These numbers just mean that the second line begins with the 18th value in the result, and the third line begins with the 35th value and so on. You can mostly ignore the numbers that appear in brackets:
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
[18] 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34
[35] 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51
[52] 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68
[69] 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85
[86] 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100: returns every integer between two integers. It is an easy way to create a sequence of numbers.If you type an incomplete command and press Enter, R will display a + prompt, which means R is waiting for you to type the rest of your command. This is because you have not closed a parenthesis or quotation, i.e. you do not have the same number of left-parentheses as right-parentheses, or the same number of opening and closing quotation marks. Either finish the command or hit Esc to start over:
> 99 +
+
+ 1
[1] 100If you type a command that R doesn’t recognize, R will return an error message. If you ever see an error message, don’t panic. R is just telling you that your computer couldn’t understand or do what you asked it to do. You can then try a different command at the next prompt:
> 100 % 2
Error: unexpected input in "100 % 2"
>Once you get the hang of the command line, you can easily do anything in R that you would do with a calculator. For example, you could do some basic arithmetic:
2 * 3
## 6
4 - 1
## 3
6 / (4 - 1)
## 2Did you notice something different about this code? I’ve left out the >’s and [1]’s. This will make the code easier to copy and paste if you want to put it in your own console.
R treats the hashtag character # in a special way; R will not run anything that follows a hashtag on a line. This makes hashtags very useful for adding comments and annotations to your code. Humans will be able to read the comments, but your computer will pass over them. The hashtag is known as the commenting symbol in R.
For the remainder of the book, we’ll use hashtags to display the output of R code. We’ll use a single hashtag to add my own comments and a double hashtag ## to display the results of code. We’ll avoid showing > and [1] unless we want you to look at them.
Exercise
That’s the basic interface for executing R code in RStudio. Think you have it? If so, try doing these simple tasks to calculate the economic order quantity step by step with the given formula (Hint: Use sqrt(x) to calculate the square root).
$$Q = \sqrt{\frac{2DK}{h}}$$
Calulate the Q where:
$D = 1000$, $K = 5$, $h = 0.25$
Solution
# Step by step
2 * 1000 * 5
## 10000
10000 / 0.25
## 40000
sqrt(40000)
## 200
# Or as an one-liner
sqrt((2 * 1000 * 5) / 0.25)
## 200Throughout the course, I’ll put exercises in chunks, like the one above. I’ll follow each exercise with a hidden model answer, like the one above.
Source
RStudio allows you to execute commands directly from the script editor by using the Ctrl + Enter shortcut (on Macs, Cmd + Return will work, too). The command on the current line in the script (indicated by the cursor) or all of the commands in the currently selected text will be sent to the console and executed when you press the shortcut.
Exercise
Write your calculations for the EOQ in the script editor and …
- … execute a single line (click the Run icon
 or hit Cmd+Return). Note that the cursor can be anywhere on the line and one does not need to highlight anything (unless you want to define a function)
or hit Cmd+Return). Note that the cursor can be anywhere on the line and one does not need to highlight anything (unless you want to define a function) - … execute multiple lines (Highlight lines with the cursor, then click Run or press Cmd+ Return
- … execute the whole script (Source icon
 or Cmd + Shift + S)
or Cmd + Shift + S)
Note that running code via source differs in a few respects from entering it at the R command line. Sourcing a file does not print command outputs. To see your output click the arrow next to the source icon ![]() and click
and click Source with Echo or use Cmd + Shift + Return.
Objects / Assignment operator
You can get now output from R simply by typing math in the console or by running a script. You generated different numbers for you to see, but it didn’t save that values anywhere in your computer’s memory. If you want to use those numbers again, you’ll have to ask your computer to save them somewhere. You can do that by creating R objects. R lets you save data by storing it inside an R object. What is an object? Just a name that you can use to call up stored data. For example, you can save data into an object like a or b. Wherever R encounters the object, it will replace it with the data saved inside.
To create an object, we need to give it a name followed by the assignment operator <- (less-than symbol, followed by a minus sign), and the value we want to give it next to it. Typing ⌥ + - (mac) will write <- in a single keystroke. R ignores spaces and line breaks and executes one complete expression at a time. But it makes the code easier for you and me to read.
Q <- 200
die <- 1:6= for assignments, but not in every context. Because of the slight differences in syntax, it is good practice to always use <- for assignments.To see what is stored in an object, just type the object’s name by itself.
Q <- 200
Q
## 200
die <- 1:6
die
## 1 2 3 4 5 6You can name an object in R almost anything you want such as x, current_temperature, or subject_id, but there are a few rules. First, a name cannot start with a number (2x is not valid, but x2 is). Second, a name cannot use some special symbols, like ^, !, $, @, +, -, /, or *. You want your object names to be explicit and not too long. There are some names that cannot be used because they are the names of fundamental functions in R (e.g. if, else, for, etc.). In general, even if it’s allowed, it’s best to not use other function names (e.g., c, T, mean, data, df, weights). It’s also best to avoid dots (.) within an object name as in my.dataset. There are many functions in R with dots in their names for historical reasons, but because dots have a special meaning in R (for methods) and other programming languages, it’s best to avoid them. It is also recommended to use nouns for object names, and verbs for function names. It’s important to be consistent in the styling of your code (where you put spaces, how you name objects, etc.). Using a consistent coding style makes your code clearer to read for your future self and your collaborators.
Exercise
Another important question here is, is R case sensitive? Is A the same as a? Figure out a way to check for yourself.
Environment
mtcars (this is a exemplary built in dataframe) to a name of your choice and take a look. Also in this pane is the History tab, where you can see all of the code executed for the session. You can also run history() to open it up. If you double-click a line or highlight a block of lines and then double-click those, you can send it to the Console (i.e., run them). The history will not be saved if we did not choose to save the environment after a session. The Build Tab will be useful later when you create your own website to hand in your code. We will come back to this shortly.Exercise
Now that we know how to put values into memory, we can do arithmetic with it and calculate EOQ again. Assign all values to R objects. Replicate the formula with those ojects, so that you just can run the line again even if another value changes.
Solution
D <- 1000
K <- 5
h <- 0.25
sqrt(2 * D * K / h)
## 200
D <- 4000
sqrt(2 * D * K / h)
## 400
Functions
R comes with many functions that you can use to do sophisticated tasks like random sampling. For example, you can round a number with the round() function, or as you have already done, calculate its square root with the sqrt() function. Using a function is pretty simple. Just write the name of the function and then the data you want the function to operate on in parentheses:
round(3.1415)
## 4
sqrt(16)
## 4The data that you pass into the function is called the function’s argument. The argument can be raw data, an R object, or even the results of another R function. In this last case, R will work from the innermost function to the outermost. Here R first looks up die, then calculates the mean of one through six, then rounds the mean:
mean(1:6)
## 3.5
die <- 1:6
mean(die)
## 3.5
round(mean(die))
## 4Many R functions take multiple arguments that help them do their job. You can give a function as many arguments as you like as long as you separate each argument with a comma.
round(3.1415)
## 3
round(3.1415, digits = 2)
## 3.14
sample(die, size = 1)
## 3With the sample function you can simulate rolling a pair of dice, if you set the argument size to the value 2. But additonally, you have to set the value replacement to TRUE. Otherwise rolling pairs would not be possible. If you want to add up the dice, you can feed your result straight into the sum function:
dice <- sample(die, size = 2, replace = TRUE)
dice
## 3 4
sum(dice)
## 7Writing your own function
You can now run this chunk again and again to simulate rolling the dice. However, this is an awkward way to work with the code. It would be easier to use your code if you wrapped it into its own function.
Every function in R has three basic parts: a name, a body of code, and a set of arguments. To make your own function, you need to replicate these parts and store them in an R object, which you can do with the function() function. To do this, call function() and follow it with a pair of braces, {}:
my_function <- function() {}function will build a function out of whatever R code you place between the braces. For example, you can turn your dice code into a function by calling:
roll <- function() {
die <- 1:6
dice <- sample(die, size = 2, replace = TRUE)
sum(dice)
}To use it, write the object’s name followed by an open and closed parenthesis:
roll()
## 6The code that you place inside your function is known as the body of the function. When you run a function in R, R will execute all of the code in the body and then return the result of the last line of code. If the last line of code doesn’t return a value, neither will your function, so you want to ensure that your final line of code returns a value.
What if you want to choose the kind of dice (number of sides of the die) each time you roll them? For that, you have to replace the object die, which indicates six faces, with another object:
roll2 <- function() {
dice <- sample(faces, size = 2, replace = TRUE)
sum(dice)
}
roll2()
## Error in sample(faces, size = 2, replace = TRUE) :
## object 'faces' not foundNow you’ll get an error when you run the function. The function needs an object faces to do its job, but there is no object named faces to be found. You can supply faces when you call roll2() if you make faces an argument of the function. To do this, put the name faces in the parentheses that follow function when you define roll2:
roll2 <- function(faces) {
dice <- sample(faces, size = 2, replace = TRUE)
sum(dice)
}
roll2(faces = 1:6)
## 7
roll2(faces = 1:10)
## 13Notice that roll2() will still give an error if you do not supply a value for the faces argument when you call roll2(). You can prevent this error by giving the faces argument a default value. To do this, set faces equal to a value when you define roll2():
roll2 <- function(faces = 1:6) {
dice <- sample(faces, size = 2, replace = TRUE)
sum(dice)
}
roll2()
## 9Exercise
What if you also want to choose the number of dice each time you roll them? Try to alter the function accordingly. Set the default value to 2 dice.
Solution
roll2 <- function(faces = 1:6, number_of_dice = 2) {
dice <- sample(x = faces, size = number_of_dice, replace = TRUE)
sum(dice)
}
roll2()
## 10
# Four Tetrahedron shaped dice (Four faces)
roll2(faces = 1:4, number_of_dice = 4)
## 11Exercise
Now transfer that approach to the EOQ formula and create a function, which calculates the EOQ and takes the variable D as an argument (set the default value of D to 1000)!
Solution
calc_EOQ <- function(D = 1000) {
K <- 5
h <- 0.25
Q <- sqrt(2*D*K/h)
Q
}
calc_EOQ()
## 200
calc_EOQ(D = 4000)
## 400Miscellaneous
Files tab has a navigable file manager, just like the file system on your operating system. It shows all the files and folders in the default workspace. The Plot tab is where graphics you create will appear. Here we can zoom, export, configure and inspect graphs/figures. Run hist(mtcars$mpg) for an example. The Packages tab shows you the packages that are installed and those that can be installed (more on this later). The Help tab allows you to search the R documentation for help and is where the help appears when you ask for it from the Console.Getting set up
It is good practice to keep a set of related data, analyses, and text self-contained in a single folder, called the working directory. It is the place from where R will be looking for and saving the files. All of the scripts within this folder can then use relative paths to files that indicate where inside the project a file is located (as opposed to absolute paths, which point to where a file is on a specific computer). Working this way makes it a lot easier to move your project around on your computer and share it with others without worrying about whether or not the underlying scripts will still work.
RStudio provides a helpful set of tools to do this through its Projects interface, which not only creates a working directory for you, but also remembers its location (allowing you to quickly navigate to it) and optionally preserves custom settings and open files to make it easier to resume work after a break. Using RStudio projects makes this easy and ensures that your working directory is set properly.
You can see your current working directory in the path given above the console or you can just run the command getwd(). If for some reason your working directory is not what it should be, you can change it in the RStudio interface by navigating in the file browser where your working directory should be, and clicking on the blue gear icon ![]()
More in the files tab and select Set As Working Directory. Alternatively you can use setwd("/path/to/working/directory") to reset your working directory. However, your scripts should not include this line because it will fail on someone else’s computer.
Using a consistent folder structure across your projects will help keep things organized, and will also make it easy to find/file things in the future. This can be especially helpful when you have multiple projects. In general, you may create directories (folders) for scripts, data, and documents.
data_raw/ & data/
Use these folders to store raw data and intermediate datasets you may create for the need of a particular analysis. For the sake of transparency and provenance, you should always keep a copy of your raw data accessible and do as much of your data cleanup and preprocessing programmatically (i.e., with scripts, rather than manually) as possible. Separating raw data from processed data is also a good idea. For example, you could have files data_raw/tree_survey.plot1.txt and ...plot2.txt kept separate from a data/tree.survey.csv file generated by the scripts/01.preprocess.tree_survey.R script.
documents/
This would be a place to keep outlines, drafts and other text.
scripts/
This would be the location to keep your R scripts for different analyses or plotting, and potentially a separate folder for your functions (more on that later).
Additional (sub)directories depending on your project needs.
Exercise
Getting set up: Create a new project, organize your working directory and save your first script:
- Download the zip file, extract the data and and choose a convenient location for it. (e.g. WIN:
C:/data-science/macOS:~/data-science)
- In RStudio under the
Filemenu, click onNew Project(or click and then choose
and then choose Existing Directory. This will be your working directory (e.g., ~/data-science). - Locate the downloaded folder and then click
Create Project.
Your working directory should now look like this (the structure was modified a little):
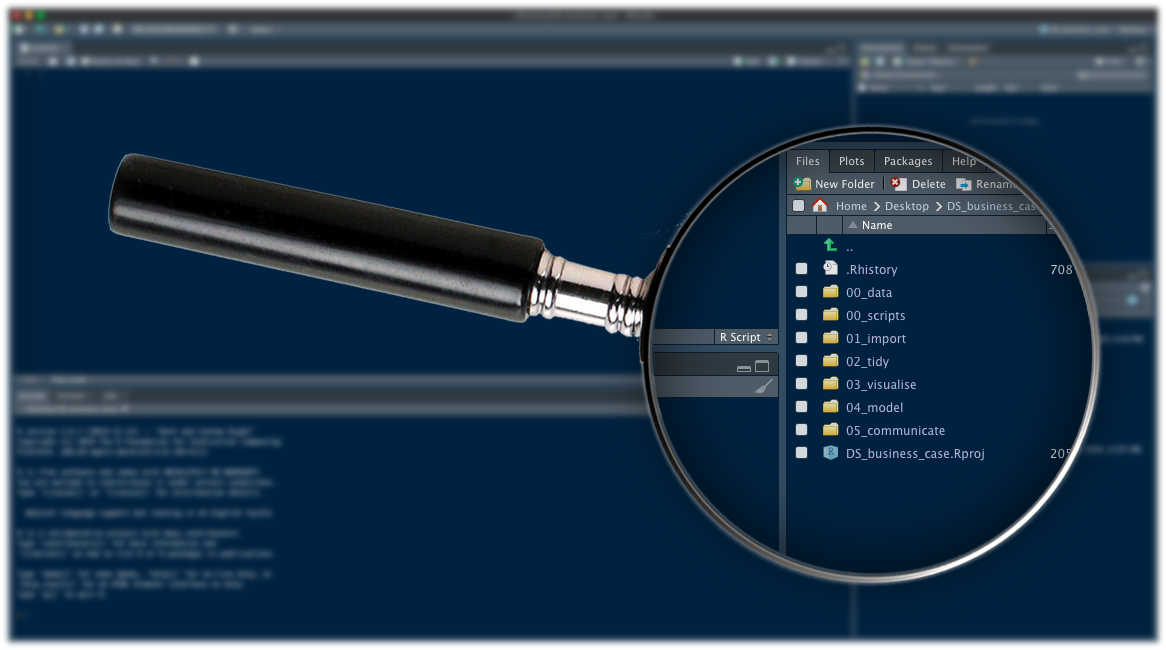
For this first project, we will just need a 01_getting_started/ folder to store our first script. We will use the other folders for when we learn how to import data such as CSV files, and for the figures that we will save.
(If it was not there already) Under the Files tab on the right of the screen, click on New Folder and create a folder named
01_getting_startedwithin your newly created working directory. (Alternatively, typedir.create("01_getting_started")at your R console.)Open your previously saved R-script and save it in this directory.
Getting help
Within RStudio
There are over 1,000 functions at the core of R, and new R functions are created all of the time. This can be a lot of material to memorize and learn! Luckily, each R function comes with its own help page, which you can access by typing the function’s name after a question mark. For example, each of these commands will open a help page. Look for the pages to appear in the Help tab of RStudio’s bottom-right pane:
?sqrt
?log10
?sample
Help pages contain useful information about what each function does. These help pages also serve as code documentation, so reading them can be bittersweet. They often seem to be written for people who already understand the function and do not need help.
Don’t let this bother you—you can gain a lot from a help page by scanning it for information that makes sense and glossing over the rest. This technique will inevitably bring you to the most helpful part of each help page: the bottom. Here, almost every help page includes some example code that puts the function in action. Running this code is a great way to learn by example.
Parts of a Help Page
Each help page is divided into sections. Which sections appear can vary from help page to help page, but you can usually expect to find these useful topics:
Description - A short summary of what the function does.
Usage - An example of how you would type the function. Each argument of the function will appear in the order R expects you to supply it (if you don’t use argument names).
Arguments - A list of each argument the function takes, what type of information R expects you to supply for the argument, and what the function will do with the information.
Details - A more in-depth description of the function and how it operates. The details section also gives the function author a chance to alert you to anything you might want to know when using the function.
Value - A description of what the function returns when you run it.
See Also - A short list of related R functions.
Examples - Example code that uses the function and is guaranteed to work. The examples section of a help page usually demonstrates a couple different ways to use a function. This helps give you an idea of what the function is capable of. You can also type example(function_name)to get examples for said function.
If you’d like to look up the help page for a function but have forgotten the function’s name, you can search by keyword. To do this, type two question marks followed by a keyword in R’s command line. R will pull up a list of links to help pages related to the keyword. You can think of this as the help page for the help page:
??logExternal to RStudio
R also comes with a super active community of users that you can turn to for help. Stack Overflow, a website that allows programmers to answer questions and users to rank answers based on helpfulness, is the best place to find an answer to your question. You can submit your own question or search through Stack Overflow’s previously answered questions related to R, because there’s a great chance that your question has already been asked and answered. You’re more likely to get a useful answer if you provide a reproducible example with your question. This means pasting in a short snippet of code that users can run to arrive at the bug or question you have in mind.
Addtionally to Stackoverflow you can just use google.com or rseek.org to search for an r related problem.
Exercise
Find a way to alter the probabilities involved in the sampling process, if your roll one die. Let’s change the probability of rolling a 6 to 50% (Because the probabilites need to sum one, the other faces have a probability of 10%). Start by opening the help page of the function sample() and take a look at the arguments.
Hint: You can create a vector with the c() function. For example:
probabilities_vector <- c(1/6, 1/6, 1/6, 1/6, 1/6, 1/6)
probabilities_vector
## 0.1666667 0.1666667 0.1666667 0.1666667 0.1666667 0.1666667Solution
roll3 <- function(faces = 1:6, number_of_dice = 1) {
dice <- sample(x = faces, size = number_of_dice,
replace = TRUE,
prob = c(0.1, 0.1, 0.1, 0.1, 0.1, 0.5))
sum(dice)
}
# You can run the function 100 times, store the results and plot a histogram to varify your function
results <- replicate(n = 100, expr = roll3(), simplify=TRUE)
hist(results)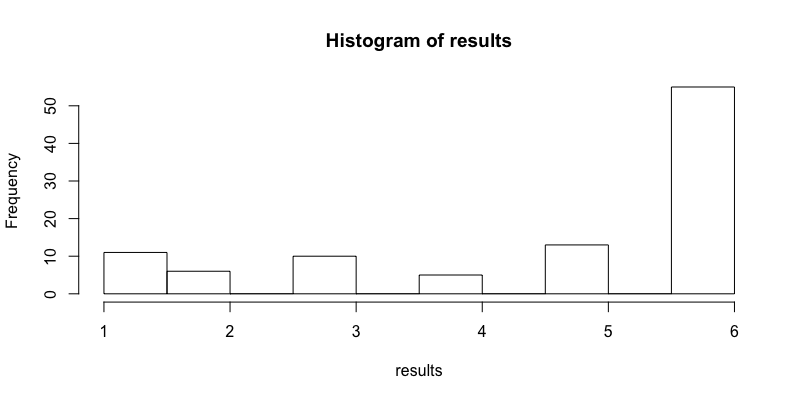
Datacamp
GitHub for journaling
We will use GitHub to store your data and hand in your assignments through your personal website (github pages). To be able to do so, complete the following steps (detailed instructions below):
- Create a free github account.
- Download, install github desktop and connect it with your account. GitHub Desktop is a graphical user interface, which allows you to sync your local code changes with your online repository.
- Windows only: Install
Git.
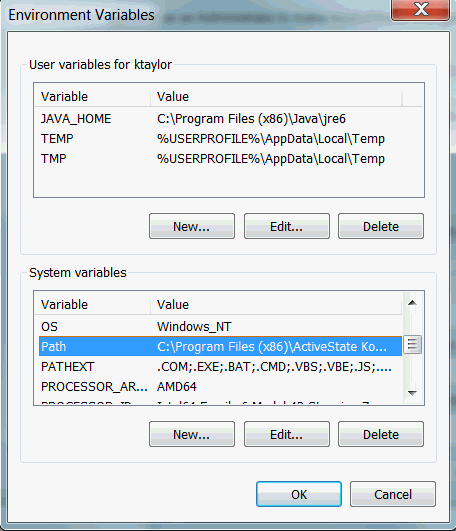
On Windows, you also have to install “Git for Windows” and set “git/cmd” and “git/bin” as system variables.
Set as system variables: System Properties (Tab: Advanced) -> Environment Variables -> System variables -> Path -> Edit -> New:
C:\Program Files\Git\cmd
C:\Program Files\Git\bin
- Accept the assignment on GitHub Classroom, create a website using R Markdown and host it on Github pages, then submit the link and your personal password to your website for the assignments. You will create the website using this template LabJournalTemplate.
To get started with this template you only have to login to github and click this invitation link:
Authorize GitHub Classroom
Accept Assignment …
You might have to refresh the page after a while. But then you’re ready to go and able to access your template by clicking on the bottom link (you don’t need to accept the invitation to TUHHStartupEngineers). That means you have all of the files you need to compile a website with RMarkdown.
If the message “owmork invited you to collaborate” pops up, press accept invitation so that all teaching assistants are able to track your progress and give feedback if needed.
Compiling on your local computer
- Make sure the
rmarkdownpackage and all it’s dependencies are installed in R-studio:
- macOS only: Install the Apple
Xcodedeveloper tools.
Option 1: Run xcode-select --install in the terminal.
You can copy the command directly in your terminal (Terminal on MacOS, Command Prompt on Windows).
Terminals/command shells only accept text commands and are a useful way to run task on a computer.
Option 2: You can install Xcode for free from the App Store.
- Windows only: Download and install the latest version of
Rtools.
- Both systems
- Open R-studio, click the packages tab in the lower right hand corner, click install packages, type in rmarkdown, make sure “install dependencies” is clicked on, then press install.
- Close R-studio.
- Open GitHub Desktop, make sure it is connected to your account and clone your repository to your computer so that you have a local copy of that data:
Sign in to GitHub.com
Continue)
Finish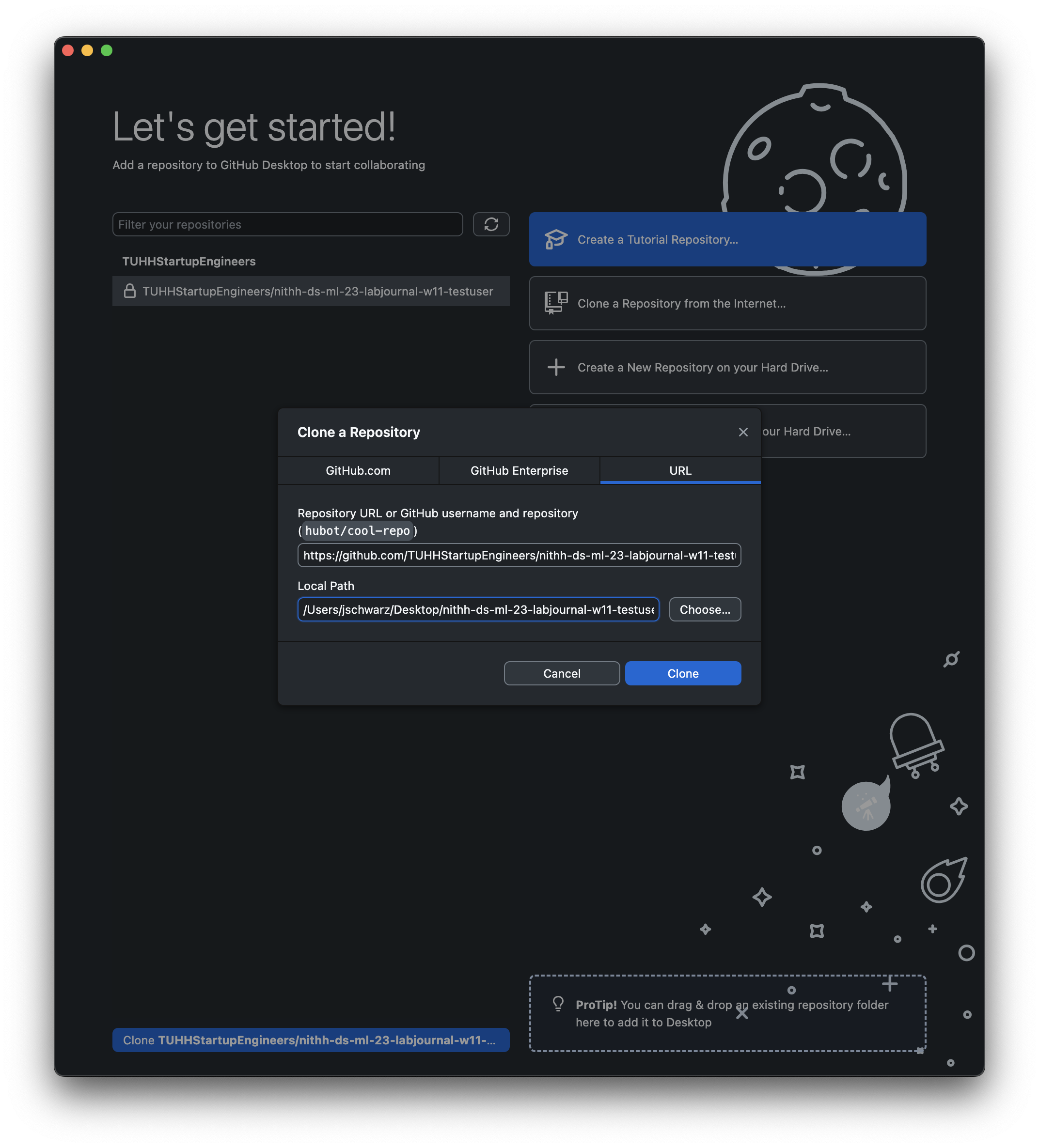
Clone a Repository from the Internet... and select your lab_journal_website repo. Choose a local path where you want to save the local copy and press Clone.Navigate to the folder you just cloned/downloaded, open the
lab_journal_website.Rprojfile. This should automatically open R-studio, and your current working environment will be inside this project. That means everything you save will be auto saved to this folder (unless you tell R-studio to save something somewhere else. Have a look at the files tab in the bottom right hand corner. Most files you click will be opened up as text files in the R-studio editor. Click the “journal.Rmd” file, replace the author name with your name and save the file again. You can freely alter the other content as well to design your website as you desire.To compile the entire website, find the build tab in the top right hand corner. You should see the option to
build website Click this. The website should be built. After the website is built, you should be able to see it in the R-studio browser (viewer pane in the bottom right corner). There is a little button
Click this. The website should be built. After the website is built, you should be able to see it in the R-studio browser (viewer pane in the bottom right corner). There is a little button  that allows you to pop the website into your default web-browser. This way you can look at the website in your browser.
Important: After compilation, all of the files for displaying your website are saved in the folder where your R project resides. When you look at these in a browser (for example, by going to the folder and dragging the index.html file into a browser), you are loading from your disk. Only you will be able to see the website, because it is on your hard-drive. You need to upload to a web server to serve the webpage on the internet.
that allows you to pop the website into your default web-browser. This way you can look at the website in your browser.
Important: After compilation, all of the files for displaying your website are saved in the folder where your R project resides. When you look at these in a browser (for example, by going to the folder and dragging the index.html file into a browser), you are loading from your disk. Only you will be able to see the website, because it is on your hard-drive. You need to upload to a web server to serve the webpage on the internet.
Serving your webpage on the internet
Every github repository has the capability of serving html files (web page files) contained in the repository, this is called github pages. How this works depends a little bit on the specific repository you are using. For this repository the webpage is served from the docs folder. The example files are set so that when you compile the example in R-studio, the output automatically goes into the docs folder. As the final step, we have to tell github to serve the html files in your docs folder as a website:
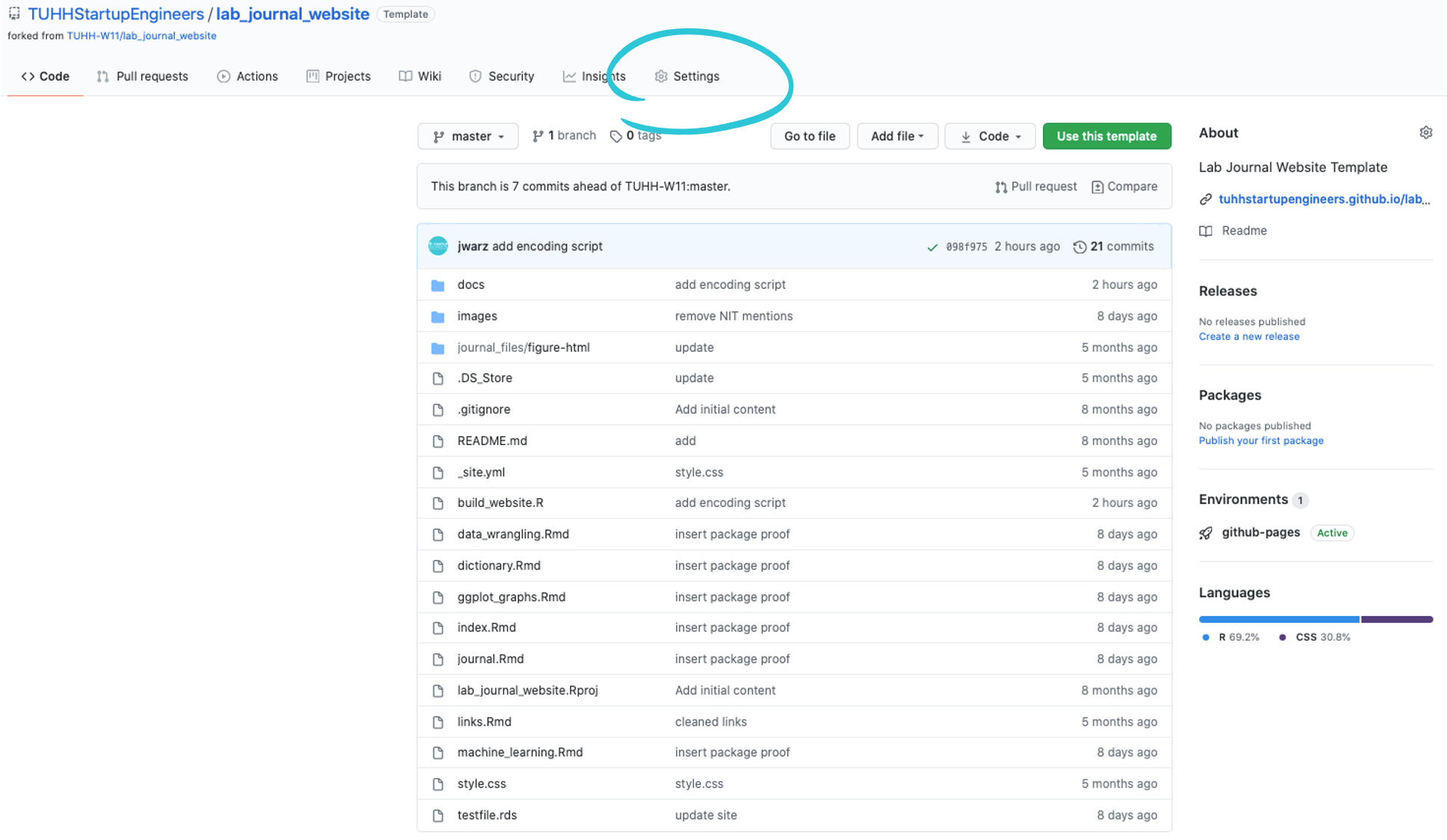
- Go to your github account, navigate to your lab_journal_website repository and click the settings button in the top right corner.
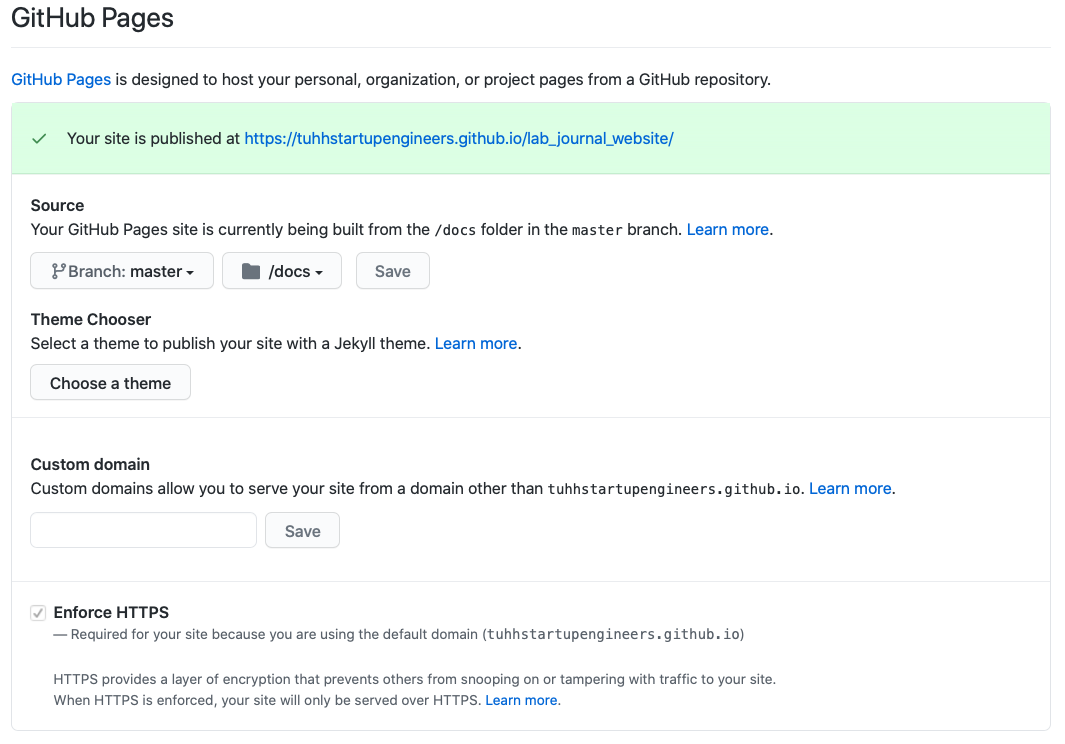
- Scroll down and select the master Branch and click the option to serve from the docs folder. Press Save. (You should see a little green message above the github pages options with a link to your new webpage).
That’s it. Now you can navigate to your new homepage via the link in the greenbox. It may take a while until you can reach it.
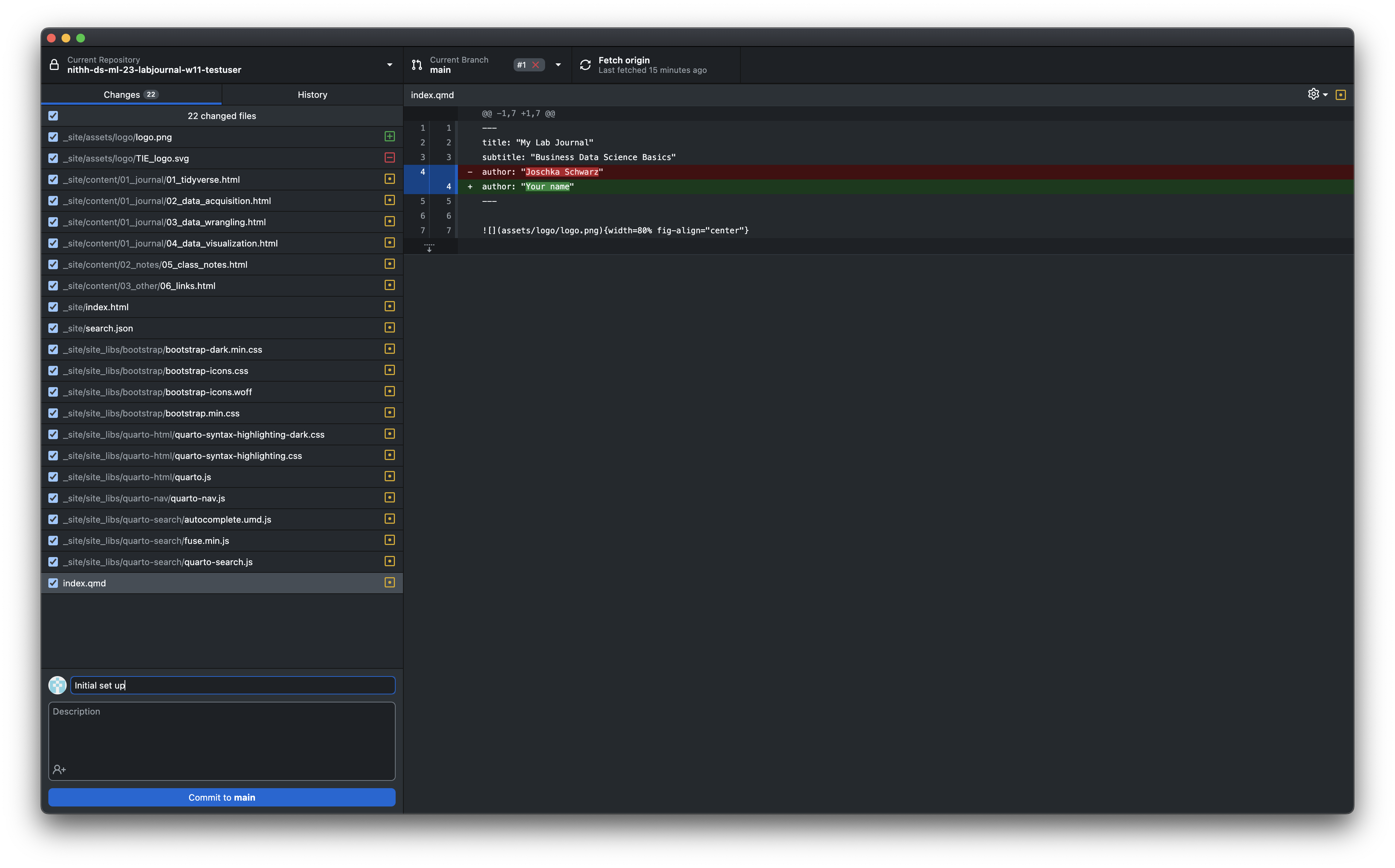
- To upload any changes to your content you have to commit your changes to github using github dektop (if you are familiar with git you are free to use another method to commit your changes). As shown in this Figure you see that GitHub Desktop detects the changes automatically after saving your files (don’t forget to build your website again after any changes. Otherwise the html files won’t be affected by any changes to your .Rmd-files). Select all of the files that you want to commit on the left panel. Write a short note to describe the changes in the box at the bottom left.
Press commit to master.
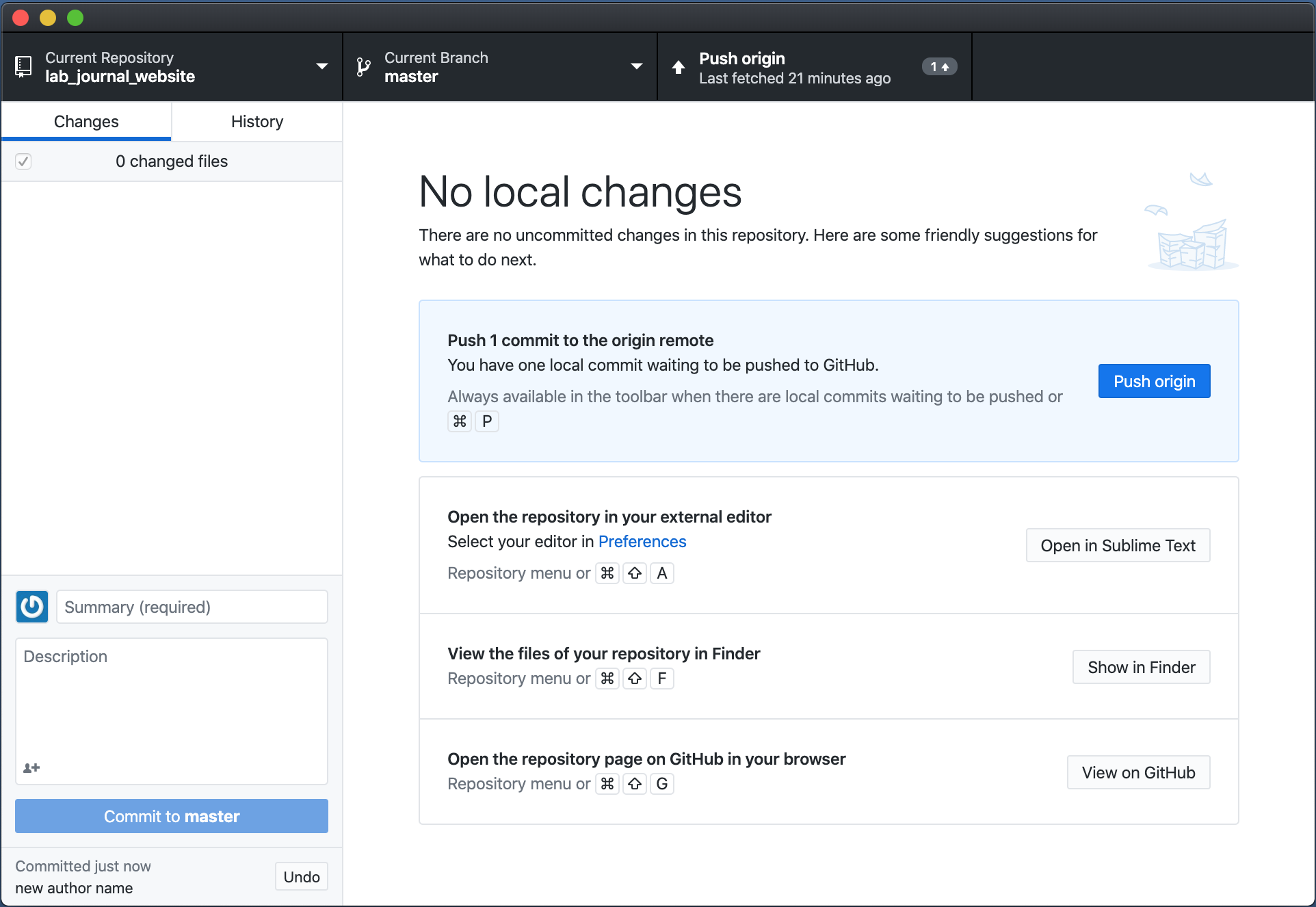
- Press
Push originand wait a couple of minutes. Your changes should now be served on your website
Now we want to go one step further and password protect you Lab Journal page (the journal.html file). We are using StatiCrypt, that uses AES-256 to encrypt your web page with your passphrase. StatiCrypt is based on the crypto-js (javascript) library, which again requires Node.js (JavaScript runtime environment that executes JavaScript code outside a web browser to produce dynamic web page content before the page is sent to the user’s web browser).
Requirements for StatiCrypt:
- Node.js
- npm (Node.js package manager)
Step 1: Installe Node.js (The Node.js installer includes the NPM package manager).
Step 2: Install StatiCrypt via npm
Run npm install -g https://github.com/TUHHStartupEngineers/staticrypt in your terminal. If you get the error: errno 13 permission denied, you don’t have the permission to install it. To resolve this error put sudo (for unix systems) in front of the command and enter your password (your password for e.g. unlocking your account on Windows/MacOS) if prompted.
Step 3: Set your password and store it in your global environment
Alternative 1
Run Sys.setenv(LAB_KEY = "Your-token") in the R console. Replace “Your-token” with your passphrase for your website. You have to repeat this step everytime you start RStudio. DO NOT USE YOUR STANDARD PASSWORDS. The lab journal password are displayed in clear text to the teaching assistants.
Alternative 2
To make this step permanent, execute the following steps:
- Create a
.Renvironfile in your project directory:file.create(".Renviron") - Open this file by clicking on it in the files browser
- Add the line
LAB_KEY = "Your-token"and replace “Your-token” with your passphrase - Save the file. If you want to access these values, you have to restart RStudio, because the
.Renvironfile is only initialized at startup. - Go to GitHub Desktop to review the changes. If the
.Renvironfile appears, right-click on it and selectIgnore File (Add to .gitignore). If it does not appear, everything is already set.
Step 4: Build your website (including password protection)
Test to check if your .Renviron variables are accessible: Run Sys.getenv("LAB_KEY") in your console. It should output your password.
Run source("build_website.R") in the R console. Your working directory needs to be set to ~/lab_journal_website/. This step also starts the process, which would have been triggered by clicking on Build Website. Additionally, it adds password protection (You can take a look at the build_website.R script to see what is going on). You have to execute step 4 everytime before you want to push changes to github / your online lab journal.
Summary for editing your webpage and serving on github:
- Open up the project file in the folder for your repo on your local computer (.rproj file).
- Edit the .rmd files in R-studio.
- Recompile website...
- ... by clicking
Build websiteto preview your changes locally. - ... by running
source("build_website.R")to add password protection (watch out that your working directory is set properly). - Send your changes back to the online github repository.
RMarkdown
Each page of your website is created by a R Markdown file (.Rmd). This is a plain text file that has the extension .Rmd. Notice that the file contains three types of content:
- An (optional) YAML header surrounded by - - - (there is no need in the beginning to alter it)
- R code chunks surrounded by
```s. These chunks can be customized withknitroptions, arguments set in the {} of a chunk header:eval = FALSEprevents running the code and include its resultsinclude = FALSEprevents code and results from appearing in the finished file. R Markdown still runs the code in the chunk, and the results can be used by other chunks.echo = FALSEprevents code, but not the results from appearing in the finished file. This is a useful way to embed figures.message = FALSEprevents messages that are generated by code from appearing in the finished file.warning = FALSEprevents warnings that are generated by code from appearing in the finished.fig.cap = "..."adds a caption to graphical results.
Example:
```{r, eval = FALSE}
numbers <- 1:1000
# This will print the first 10 elements of the vector numbers
numbers[1:10]
# This will plot a histogram of 100 random elements of the vector numbers
hist(sample(numbers, 100, replace = T))
```Check for yourself what happens, if you set eval to TRUE.
- text mixed with simple text formatting
See the R Markdown Cheat Sheet for further information regarding the markdown syntax:
Submission
Submit your lab journal URL and password via the following form: